- Methodology article
- Open access
- Published:
Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design
BMC Biotechnology volume 16, Article number: 54 (2016)
Abstract
Background
Highly multiplexed assays for quantitation of RNA transcripts are being used in many areas of biology and medicine. Using data generated by these transcriptomic assays requires measurement assurance with appropriate controls. Methods to prototype and evaluate multiple RNA controls were developed as part of the External RNA Controls Consortium (ERCC) assessment process. These approaches included a modified Latin square design to provide a broad dynamic range of relative abundance with known differences between four complex pools of ERCC RNA transcripts spiked into a human liver total RNA background.
Results
ERCC pools were analyzed on four different microarray platforms: Agilent 1- and 2-color, Illumina bead, and NIAID lab-made spotted microarrays; and two different second-generation sequencing platforms: the Life Technologies 5500xl and the Illumina HiSeq 2500. Individual ERCC controls were assessed for reproducible performance in signal response to concentration among the platforms. Most demonstrated linear behavior if they were not located near one of the extremes of the dynamic range. Performance issues with any individual ERCC transcript could be attributed to detection limitations, platform-specific target probe issues, or potential mixing errors. Collectively, these pools of spike-in RNA controls were evaluated for suitability as surrogates for endogenous transcripts to interrogate the performance of the RNA measurement process of each platform. The controls were useful for establishing the dynamic range of the assay, as well as delineating the useable region of that range where differential expression measurements, expressed as ratios, would be expected to be accurate.
Conclusions
The modified Latin square design presented here uses a composite testing scheme for the evaluation of multiple performance characteristics: linear performance of individual controls, signal response within dynamic range pools of controls, and ratio detection between pairs of dynamic range pools. This compact design provides an economical sample format for the evaluation of multiple external RNA controls within a single experiment per platform. These results indicate that well-designed pools of RNA controls, spiked into samples, provide measurement assurance for endogenous gene expression studies.
Background
In 2003, the National Institute of Standards and Technology (NIST) hosted a meeting to discuss the need for a universal RNA reference material, which could be used for gene expression profiling assays [1]. As a result of this effort, the External RNA Controls Consortium (ERCC) was formed, of which NIST is a founding member and host. The ERCC assembled a sequence library of 176 DNA sequences that could be transcribed into RNA to serve as controls in systems used to measure gene expression [2, 3]. These controls were cataloged as ERCC-00001 through ERCC-00176, and are collectively referred to as ERCC controls in this manuscript. These were evaluated and a subset was selected for dissemination as a standard. A set of 96 controls are now available as a set of sequence-certified DNA plasmids, NIST Standard Reference Material (SRM) 2374 [4].
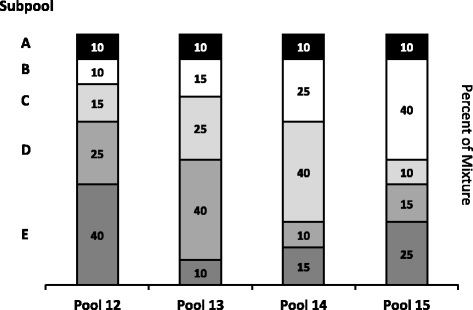
In the final phase of evaluation, an experimental design for assessing the combined performance of ERCC controls prepared as complex RNA pools was used. Each ERCC subpool was designed to have a 220 dynamic range of abundance of controls, and particular controls in the different pools were present in different abundances according to a modified Latin square design. This design provides known relative differences between the pools across a large dynamic range of abundance (Fig. 1). With this design, individual ERCC controls were assessed for their signal response to 1.5-, 2.5-, and 4-fold increases in concentration. Pairwise comparisons of these pools also provide for an assessment of ratio-based performance as a function of dynamic range. Initially assessed with three different microarray platforms, these same pools were subsequently measured by RNA sequencing (RNA-Seq) with two second-generation (NGS) sequencing platforms. The data from these two sets of experiments, corresponding to the 96 controls of the SRM, are presented here.
Modified Latin square pool design. Five subpools of ERCC controls, each containing a unique set of transcripts at different target relative abundance (see Table 1), were combined in varying proportions. Subpools A, B, C, D, and E are shaded, black, white, light grey, medium grey, and dark grey, respectively. Subpool A is present as a constant 10 % component of each pool, the modification. Subpools B-E are mixed using a Latin square of proportions 10, 15, 25, and 40 %
Methods
Pool design
The ERCC controls were distributed into 5 subpools (A-E), each containing a unique set of controls (see Table 1). These subpools were prepared at AIBioTech (formerly CBI Services, Richmond, VA) to ERCC specifications. This design results in the relative abundance within each subpool covering a dynamic range of 220. Subpools A-E were then mixed by volume in a modified Latin square design to create 4 different pools (see Fig. 1 and Table 1). Subpools B-E have different relative abundances between the four pools (in a Latin square design), while subpool A is held at a constant proportion (the “modification”). In addition, the ERCC controls in subpools B-E participate in 6 pairwise comparisons between pools to produce ratios of 4-, 2.7-, 2.5-, 1.7-, 1.6-, and 1.5-to-1 (Fig. 1 and Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5 and Additional file 6: Figure S6). The ERCC controls in subpool A are always present at 10 % in any of the pools, and create the 1-to-1 component in any of the 6 possible pairwise comparisons. These pools were designated as Pools 12, 13, 14, and 15 in the set of pools developed for ERCC testing [2]. Each pool was spiked into a common “background” of human liver total RNA (Ambion) to create 4 corresponding samples. Each microarray test site determined the relative amount of spike-in pools to add to the background. Agilent, Illumina, and NIAID used 0.144, 0.25, and 0.265 % (wt/wt) of ERCC pool per total liver RNA, respectively. For the sequencing test sites total RNA samples were spiked at NIST at 0.3 % (wt/wt) and then sequenced by NIST and Illumina.
The ERCC molecules used in these pools were prepared by in vitro transcription of polymerase chain reaction (PCR) products representing candidate sequences prior to the release of NIST SRM 2374. The plasmids were designed to produce either “sense” or “antisense” RNA controls [4]. In this study, seven of these ERCC transcripts were determined to be antisense using a stranded RNA-Seq protocol (see Table 1) and were excluded from further data analysis, because the microarrays were designed to detect sense RNA controls.
Microarray measurements
Samples were measured at each test site using the following methods.
The NIAID in-house spotted microarrays contain long (70-mer) oligonucleotides designed to hybridize the ERCC transcripts printed on epoxy-coated glass slides (Corning) in quadruplicate using an OmniGrid robot (Genomic Solutions) with 16 SMP3 print tips (Telechem). RNA was reverse transcribed using Oligo dT primer (12–20 mer) mix (Invitrogen) and Superscript II reverse transcriptase (Invitrogen). Fluorescent Cy-Dye-dUTP (GE) nucleotide was incorporated into first-strand cDNA during the reverse transcription. After degradation of the mRNA template strand, labeled single-stranded cDNA target was purified using Vivaspin 500 (10 K, Millipore). Hybridization was performed at 45 C°, for 16 h on a MAUI hybridization station. The arrays were washed twice in 1X SSC and 0.05 % SDS and twice in 0.1X SSC, then air dried. Microarrays were scanned on GenePix 4000B (Axon) at 10 μm resolution. GenePix Pro software was used for image analysis. Median pixel intensity (no background subtraction) was taken for each of the 4 replicate spots, the median of these four values was taken to represent the data.
The Agilent microarrays (8x60K Agilent G3 8-pack format with the Design ID 022439) contain 60-mer oligonucleotide probes synthesized in situ onto slides using a proprietary non-contact industrial inkjet printing process. Labeled cRNA for both the one-color and two-color microarray experiments was prepared using the Agilent Low Input Quick Amp Labeling Kit, Two-Color (5190-2306). RNA was reverse transcribed using AffinityScript RT, Oligo(dT) Promoter Primer, and T7 RNA Polymerase. Fluorescent Cy-Dye-dCTP nucleotide was incorporated during cRNA synthesis and amplification. Microarrays were hybridized at 65 °C for 17 h. All microarrays were scanned in one batch in random order using default settings for Agilent C Scanner using a single pass over the scan area at a resolution of 3 μm and a 20-bit scan type. Data was extracted with Agilent Feature Extraction Software (ver. 10.7.3.1) using the default settings for either the one-color protocol or the two-color protocol.
The Illumina Human-6 Expression BeadChips contain 50-mer oligonucleotide probes with a 29-mer address sequences attached to beads held in etched microwells. RNA was reverse transcribed using a T7 Oligo(dT) primer containing a T7 promoter sequence. Biotinylated cRNA was prepared using the Illumina TotalPrep RNA Amplification Kit (Ambion). BeadChips were hybridized at 58 °C for 14–20 h, washed, and labeled with streptavidin-Cy3. BeadChips were scanned with the Illumina iScan System. Intensity values are determined for every bead and summarized for each bead type. For more details refer to the Whole-Genome Gene Expression Direct Hybridization Assay Guide (Illumina, part no. 11322355).
RNA sequencing measurements
NIST prepared samples of spiked liver total RNA for sequencing analysis with the 5500xl at NIST and the HiSeq 2500 at Illumina. Prior to library preparation samples were depleted of ribosomal RNA. The 5500xl experiment produced an average of 23,866,495 single-ended reads (75 base) per sample and the HiSeq 2500 experiment yielded an average of 48,168,710 paired-end reads (2 x 75 base). For both platforms sequence reads were aligned against a reference sequence consisting of the human genome (hg19) and the ERCC transcript sequences of SRM 2374 (Note: ERCC-00114 is not part of the SRM and not included as part of the reference transcriptome). Alignment and quantification of sequence reads to obtain per transcript counts was performed with the LifeScope bioinformatic analysis suite (Life Technologies) for 5500xl data and the Tophat-Cufflinks suite was used for HiSeq 2500 data [5, 6].
Results and discussion
For each of the platforms, if the ERCC spike-in pools are added to the background RNA in the proper proportion, then the 220 range of relative abundance will cover the distribution of the endogenous transcript signals. In the first set of experiments, each microarray platform provider empirically determined in pilot studies its chosen spike-in proportion to add to the total RNA background (not shown). Agilent used 0.144 % (wt/wt) for both one-color and two-color arrays, and Illumina and NIAID used 0.25, and 0.265 %, respectively. For the RNA-Seq experiments, ERCC pools were added to the background at NIST at 0.3 % and shared with the Illumina site. The LifeTech 5500xl and Illumina HiSeq measurements were performed at NIST and Illumina, respectively. The distribution of ERCC signals relative to the endogenous liver background transcripts are shown for all platforms in Table 2. For all sites, the dynamic range of the signals from the controls matched the range of signal expression from the endogenous genes of the liver background. This supports the use of these signals to derive metrics useful for characterizing each measurement system (see Additional files 7, 8, 9, 10, 11 and 12 for ERCC data).
Dose-response and outlier detection
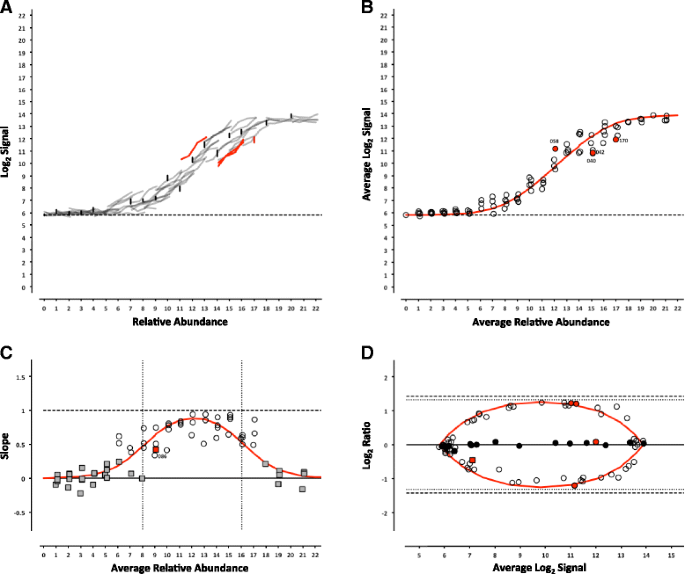
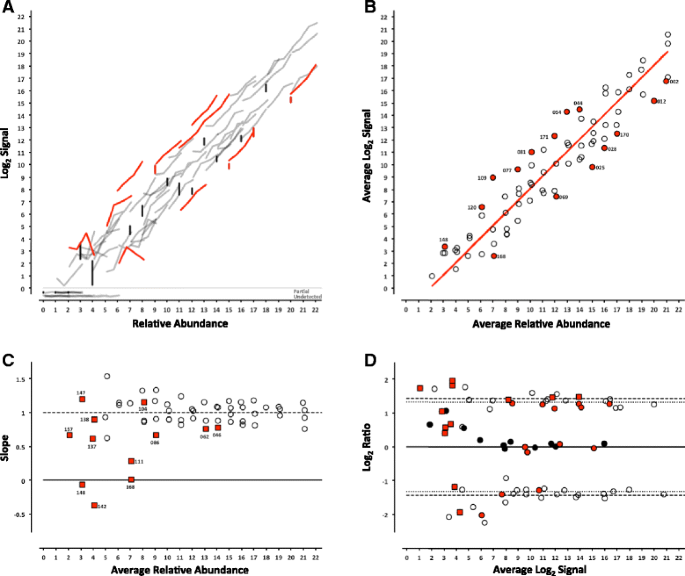
For each platform, we can determine whether the analytical signal (fluorescence intensity in microarrays or length normalized counts in sequencing) changes with the concentration of an analyte (the ERCC being measured). For each control, the signal from each pool can be plotted against the corresponding relative abundance (Table 1), producing a collection of dose-response curves representing each individual ERCC in the study (Figs. 2, 3, 4, 5, 6 and 7, panel a). ERCC controls that were missing data for one or more concentrations in the RNA-Seq experiments were flagged as partially detected or undetected, and excluded from further analysis (Figs. 6 and 7, panel a). The mid-point of each ERCC dose-response curve (average signal versus average relative abundance from the Latin square) was used to assess whether any particular ERCC was an outlier relative to the entire set of controls. The data were fit to an appropriate model for each platform (Figs. 2, 3, 4, 5, 6 and 7, panel b).
ERCC signal response as a function of relative abundance in each of the four pools on the Illumina microarray platform. In Panel a, each line represents an individual ERCC control, where grey = titrated, black = 1-to-1, red = outlier, and dashed-line = background (average ERCC-00073). In Panel b, the centroid of each ERCC control is plotted, where the red line corresponds to the fitted Langmuir model, open circles = within 99 % CI, red circles = outliers, and dashed-line = background. In Panel c, the slope of each ERCC is plotted, where the red line corresponds to expected slope (first derivative of the Langmuir model), the vertical dotted lines correspond to the margins of the linear region (inflection points of the first derivative of the Langmuir model), the open circles = monotonic ERCC controls (ρ = 1), grey squares = non-monotonic, and red = outliers. Numbers adjacent to outliers in Panels b and c correspond to the last three digits of the Control ID in Table 3. In Panel d, each ERCC control is represented on the Bland-Altman plot of Pool 12 vs Pool 14, where the red line corresponds to the ratio versus average intensity derived from the fitted Langmuir model, with outliers coded as in Panels b and c above
ERCC signal response as a function of relative abundance in each of the four pools on the NIAID microarray platform. See Fig. 2 legend
ERCC signal response as a function of relative abundance in each of the four pools on the Agilent 1-color microarray platform. See Fig. 2 legend
ERCC signal response as a function of relative abundance in each of the four pools on the Agilent 2-color microarray platform. See Fig. 2 legend
ERCC signal response as a function of relative abundance in each of the four pools on the LifeTech NGS platform. In Panel a, each line represents an individual ERCC control, where grey = titrated, black = 1-to-1, and red = outlier. Partially detected and undetected ERCC controls are included at the bottom to indicate their targeted relative abundance. In Panel b, the centroid of each ERCC control is plotted, where the red line corresponds to the linear fitted model, open circles = within 99 % CI, and red circles = outliers. In Panel c, the slope of each ERCC is plotted, where the open circles = monotonic ERCC controls (ρ = 1), grey squares = non-monotonic, and red = outliers. Numbers adjacent to outliers in Panels b and c correspond to the last three digits of the Control ID in Table 3. In Panel d, each ERCC control is represented on the Bland-Altman plot of Pool 12 vs Pool 14, with outliers coded as in Panels b and c above
ERCC signal response as a function of relative abundance in each of the four pools on the Illumina NGS platform. See Fig. 6 legend
For the microarray experiments, a model using the Langmuir isotherm was used [7, 8]. The dissociation constant, K d , was determined by fitting the data as follows:
Where the maximal intensity of a feature at saturation, I max , and the background, bg, are experimentally derived from the average of the most abundant ERCC in each of the 4 pools and ERCC-00073, a component omitted from the pools, respectively. For the RNA-Seq experiments, a linear fit with a slope of 1 and fitted y-intercept was used as the model. For either model, ERCC controls outside the 99 % confidence interval (CI) were flagged as outliers (Figs. 2, 3, 4, 5, 6 and 7, panel b) and compared across platforms to identify any ERCC-specific anomalies (Table 3).
With the exception of the ERCC controls in the 1-to-1 subpool, the signal for each control should follow a strictly increasing monotonic function determined by the pool fraction of the Latin square design, 10 % < 15 % < 25 % < 40 %, (see Fig. 1). This monotonicity was assessed with Spearman’s rho, ρ, where ERCC controls with ρ < 1 were identified for comparison across platforms.
In addition, the slope of each individual ERCC dose-response curve can be calculated and plotted as a function of the relative abundance, where the slope (m = 1) corresponds to an ideal dose-response. For the microarray data, the first derivative of the Langmuir function also provides us with a model of the expected slope and the inflection points allow us to demarcate a region of the dynamic range where we should expect a linear response (Figs. 2, 3, 4, 5, 6 and 7, panel c) [9]. Non-monotonic ERCC controls that fall within that portion of the dynamic range were also identified as outliers. For the RNA-Seq data, all non-monotonic ERCC controls are flagged as outliers (Figs. 6 and 7, panel c). One control, ERCC-00113, was an outlier on all platforms, with ρ = −0.2 for each. Closer inspection of the monotonic trend indicated that the least abundant target feature produced the highest signal in each case. This ERCC was more consistent with membership in subpool C, indicating a likely error in the preparation of the subpools. Therefore, Figs. 2, 3, 4, 5, 6 and 7 include this ERCC plotted as a component of subpool C.
Table 3 includes all ERCC identified as outliers by the two criteria above and highlighted in Figs. 2, 3, 4, 5, 6 and 7 and specific controls discussed below are indicated with an asterisk (*). The majority of non-monotonic ERCC controls in the microarray experiments occurred below the lower inflection point on the slope plots and those flagged for non-detection in the RNA-Seq experiments also appear in the lower range of the signal response curves. For these ERCC controls, it is difficult to assess performance beyond their utility for defining the lower limits of the linear range, so these are not included in the outlier table.
There were nine controls that were outliers on at least one platform for each criteria. Six of those were outliers for both criteria on the same platform: ERCC-00156 on LifeTech; ERCC-00131, ERCC-00134 and ERCC-00143 on AGL-1; ERCC-00148 on AGL-1 and ILM HiSeq; and ERCC-00168 on AGL-2 and ILM HiSeq. All of these controls performed well on the majority of platforms.
Fifteen ERCC controls were non-monotonic only. ERCC-00046 and ERCC-00062 were the most highly abundant outliers in this class. In both cases, the two lowest concentrations for each control produced nearly identical values where the lowest concentration is slightly higher. With the exception of ERCC-00138, all of these controls performed well on the majority of platforms.
There are 26 ERCC controls that appear to be outliers with respect to the overall dose-response model that are still monotonic. For example, ERCC-00058 was the only control to be determined a response curve outlier on all microarray platforms and one RNA-Seq platform, however the observed slope on all platforms tested was greater than 0.9. ERCC-00170 was also flagged on every platform except the NIAID microarray, but was not evaluated for montonicity because it is in the 1-to-1 subpool.
Some of these results may be attributable to difficulties with accurately preparing large dynamic range pools with multiple controls, so that the actual concentration is different than the nominal abundance. The linear signal responses observed within the Latin square design indicate that the proper combination of subpools A-E was achieved. Some of these outliers might also be the result of an RNA processing bias that may be analyte specific and proportional to abundance, for example poly-A enrichment [10].
Intensity-dependent differential expression
For microarray data, an intensity-dependent bias is often visualized using an MA-plot; where M is the log2 transformation of the ratio of red and green fluorescence intensities in 2-channel data, and A is the log2 transformation of the average of the two [11]. This view has also been applied to two-condition single channel data, where M becomes the ratio of two different conditions, which is also referred to as a ratio-intensity plot (RI-plot) [12]. These comparative visualizations have been extended to sequencing data in the form of RA-plots, where the ratios and averages of integer count data form a characteristic pattern at the lower end of the signal range [13]. Each of these visualizations is a variation of a Bland-Altman plot (or difference plot), which is used here to visualize the ability to detect the nominal differences between two measurements [14]. A Bland-Altman plot of the ERCC components can be generated for any pairwise combination of Pools 12–15. One possible pairwise comparison, which produces fold-changes of 2.5 and 2.7 in both “up” and “down” directions (see Fig. 1) is shown in Figs. 2, 3, 4, 5, 6 and 7, panel d. Additional pairwise comparisons are shown in Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5 and Additional file 6: Figure S6.
For the microarray platforms, the discrimination between the target ratios is optimal near the middle of their dynamic range, and the ratios are “compressed” at both the lower and upper extremes. This constraint upon log2 ratios has been previously described [15]. The ratios converge towards unity at lower end due to background noise, which is additive, and contributes to both samples being compared. A similar compression is seen at high signal, where saturation dominates. We can also use Equation 1 to derive the expected intensity ratios and average intensities for any fold-change of relative abundance. These fitted curves are also shown in Figs. 2, 3, 4 and 5, panel d.
Signals in RNA-Seq are not subject to saturation (though high abundance transcripts can dominate the counting, and “crowd out” signals from lower abundance controls). As a consequence, the ratios do not compress at the upper end of the dynamic range. The RNA-Seq signals in this dataset are derived from counting technical replicates, where the variation can be characterized by a Poisson distribution [16]. In this case, “shot noise” dominates the signal at the low end, where counts might be added to either sample, and the ratios may deviate from target values in either direction (Figs. 6 and 7, panel d).
Conclusions
The modified Latin square design provided for simultaneous evaluation of multiple controls with a minimal number of samples. While each individual ERCC control was only tested over a small range of relative abundance, up to 4-fold for the ERCC controls tested at multiple ratios and a single relative abundance value for the 1-to-1 components, in aggregate, they describe the overall measurement behavior of a platform.
The spread of the data indicates that differences in signals observed between different RNA species within the same sample may not accurately reflect the relative abundance between different RNA components of the same sample. Some of this dispersion may be due to the complexity of the pools used in these experiments where the distribution of target abundances described in Table 1 may not have been attained. For microarrays, probe designs for each ERCC target may also introduce some variability in signal between different ERCC controls at the same relative abundance. For RNA-Seq, a non-uniform distribution of reads along different control sequences may also contribute to the variability [17].
The ERCC controls did demonstrate that there is a linear region of the dynamic range of each platform where changes in abundance of a particular RNA transcript can produce a proportional change in signal. In this region, the ratios obtained with each platform approach the target ratios of the modified Latin square design. As a consequence, comparisons between samples for any particular RNA species can be expected to be accurate with respect to ratio-based measurements if they fall within this region. A pair of complex mixtures of RNA controls derived from NIST SRM 2374 designed to provide a set of ratios across a similar dynamic range is commercially available (Ambion™ ERCC ExFold RNA Spike-In Mixes). NIST has developed an R-based tool, the erccdashboard, to provide metrics and visualizations for these controls [18].
The ERCC RNA controls demonstrated utility in four different gene expression microarray platforms and two RNA-Seq platforms. Performance issues with any individual ERCC could be attributed to detection limitations or a target probe issue for particular platforms. The spike-in RNA controls were useful for establishing the dynamic range of relative abundance for a platform as well as delineating a reliable region where ratios can be measured accurately.
The composite testing scheme used in this study demonstrated that using well-designed pools of RNA controls provides measurement assurance for endogenous gene expression studies. Pools of RNA controls from this study have been used as spike-ins for RNA-Seq experiments [19], and commercially available versions of these controls have been used for their intended purpose as quality controls [20–23]. These controls have also proven useful in product and method development due to their certified sequences and known concentrations [24–32]. Recently, they have become important in comparing transcriptomes between cell types in immunology [20, 32, 33], agriculture [34, 35], and other biology studies [21, 36–38], as well as key to understanding and accounting for the technical noise in single-cell sequencing studies [39–42].
Abbreviations
AGL-1, agilent 1-color; AGL-2, agilent 2-color; ERCC, External RNA Controls Consortium; ILM-bead, illumina bead array; ILM-ngs, illumina next-generation sequencing; LifeTech, Life Technologies; NGS, next-generation sequencing; NIAID, National Institute of Allergy and Infectious Diseases; NIST, National Institute of Standards and Technology; PCR, polymerase chain reaction; RNA-Seq, RNA sequencing; SRM, standard reference material
References
Cronin M, Ghosh K, Sistare F, Quackenbush J, Vilker V, O'Connell C. Universal RNA reference materials for gene expression. Clin Chem. 2004;50(8):1464–71.
External RNA Controls Consortium. Proposed methods for testing and selecting the ERCC external RNA controls. BMC Genomics. 2005;6:150.
External RNA Controls Consortium. External RNA Controls Consortium: a progress report. Nat Methods. 2005;2(10):731–4.
NIST SRM 2374 Certificate of Analysis (2013); https://www-s.nist.gov/srmors/certificates/2374.pdf.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78.
Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22.
Halperin A, Buhot A, Zhulina EB. On the hybridization isotherms of DNA microarrays: the Langmuir model and its extensions. J Phys Condens Matter. 2006;18:S463–90.
Held GA, Grinstein G, Tu Y. Relationship between gene expression and observed intensities in DNA microarrays--a modeling study. Nucleic Acids Res. 2006;34(9):e70.
Sebaugh JL, McCray PD. Defining the linear portion of a sigmoid-shaped curve: bend points. Pharmaceut Statist. 2003;2:167–74.
Qing T, Yu Y, Du T, Shi L. mRNA enrichment protocols determine the quantification characteristics of external RNA spike-in controls in RNA-Seq studies. Sci China Life Sci. 2013;56(2):134–42.
Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002;12:111–39.
Pine PS, Rosenzweig BA, Thompson KL. An adaptable method using human mixed tissue ratiometric controls for benchmarking performance on gene expression microarrays in clinical laboratories. BMC Biotechnol. 2011;11:38.
Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci U S A. 2012;109(6):E317–25.
Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8(2):135–60.
Sharov V, Kwong KY, Frank B, Chen E, Hasseman J, Gaspard R, et al. The limits of log-ratios. BMC Biotechnol. 2004;4:3.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8.
Finotello F, Lavezzo E, Bianco L, Barzon L, Mazzon P, Fontana P, et al. Reducing bias in RNA sequencing data : a novel approach to compute counts. BMC Bioinformatics. 2014;15 Suppl 1:S7.
Munro SA, Lund SP, Pine PS, Binder H, Clevert DA, Conesa A, et al. Assessing technical performance in differential gene expression experiments with external spike-in RNA control ratio mixtures. Nat Commun. 2014;5:5125.
Jiang L, Schlesinger F, Davis CA, Zhang Y, Li R, Salit M, et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21(9):1543–51.
Fritz EL, Rosenberg BR, Lay K, Mihailović A, Tuschl T, Papavasiliou FN. A comprehensive analysis of the effects of the deaminase AID on the transcriptome and methylome of activated B cells. Nat Immunol. 2013;14:749–55.
Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:1–11.
Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2012;2:666–73.
Fu GK, Xu W, Wilhelmy J, Mindrinos MN, Davis RW, Xiao W, et al. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc Natl Acad Sci. 2014;111:1891–6.
Devonshire AS, Sanders R, Wilkes TM, Taylor MS, Foy CA, Huggett JF. Application of next generation qPCR and sequencing platforms to mRNA biomarker analysis. Methods. 2013;59:89–100.
Kralj JG, Salit ML. Characterization of in vitro transcription amplification linearity and variability in the low copy number regime using External RNA Control Consortium (ERCC) spike-ins. Anal Bioanal Chem. 2013;405:315–20.
Sanders R, Mason DJ, Foy CA, Huggett JF. Evaluation of Digital PCR for Absolute RNA Quantification. PLoS One. 2013;8:e75296.
Devonshire AS, Elaswarapu R, Foy CA. Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC Genomics. 2010;11:662.
Mercer TR, Clark MB, Crawford J, Brunck ME, Gerhardt DJ, Taft RJ, et al. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat Protoc. 2014;9:989–1009.
Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108.
Law CW, Chen Y, Shi W. Smyth GK: voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Zhu Y, Li M, Sousa AM, Šestan N. XSAnno: a framework for building ortholog models in cross-species transcriptome comparisons. BMC Genomics. 2014;15:343.
Mohammadi P, di Iulio J, Muñoz M, Martinez R, Bartha I, Cavassini M, et al. Dynamics of HIV Latency and Reactivation in a Primary CD4+ T Cell Model. PLoS Pathog. 2014;10:e1004156.
Shin H, Shannon CP, Fishbane N, Ruan J, Zhou M, Balshaw R, et al. Variation in RNA-Seq Transcriptome Profiles of Peripheral Whole Blood from Healthy Individuals with and without Globin Depletion. PLoS One. 2014;9:e91041.
MacGregor DR, Gould P, Foreman J, Griffiths J, Bird S, Page R, et al. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell. 2013;25:4391–404.
Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, et al. Insights into the Maize Pan-Genome and Pan-Transcriptome. Plant Cell. 2014;26:121–35.
Sheean ME, McShane E, Cheret C, Walcher J, Müller T, Wulf-Goldenberg A, et al. Activation of MAPK overrides the termination of myelin growth and replaces Nrg1/ErbB3 signals during Schwann cell development and myelination. Genes Dev. 2014;28:290–303.
Malone JH, Cho DY, Mattiuzzo NR, Artieri CG, Jiang L, Dale RK, et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13:R28.
Jagya N, Varma SP, Thakral D, Joshi P, Durgapal H, Panda SK. RNA-Seq based transcriptome analysis of hepatitis e virus (HEV) and hepatitis B virus (HBV) replicon transfected Huh-7 cells. PLoS One. 2014;9(2):e87835.
Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–5.
Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–60.
Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343(6172):776–9.
Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–40.
Acknowledgements
Not applicable.
Funding
This research was supported in part by the Intramural Research Program of the NIH, NIAID.
Availability of data and material
The datasets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
ABL, JL, TGM, and MS designed the study. JM, SMJ-H, and MS developed the reference samples. SAM, JRP, JM, ABL, JL, TGM, and QS acquired and processed the data. All authors participated in the preliminary analysis and interpretation. PSP developed metrics and visualizations and drafted the manuscript. All authors participated in the revision process and provided final approval.
Competing interests
JL is employed by Illumina Inc., manufacturer of one of the microarray platforms and one of the sequencing platforms used in this study. ABL is employed by Agilent Inc., manufacturer of one of the microarray platforms used in this study. These authors provided data. All data analysis was performed by NIST.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Disclaimer
Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology (NIST), nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Figure S1.
Bland-Altman plot of each pair-wise pool comparison using the Illumina microarray platform. Symbols correspond to pools A-E (see Fig. 1). Filled circles = A, open circles = B, open diamonds = C, open triangles = D, and open squares = E. The red line corresponds to the ratio versus average intensity derived from the fitted Langmuir model. (PDF 249 kb)
Additional file 2: Figure S2.
Bland-Altman plot of each pair-wise pool comparison using the NIAID microarray platform. See Additional file 1: Figure S1 legend. (PDF 262 kb)
Additional file 3: Figure S3.
Bland-Altman plot of each pair-wise pool comparison using the Agilent 1-color microarray platform. See Additional file 1: Figure S1 legend. (PDF 279 kb)
Additional file 4: Figure S4.
Bland-Altman plot of each pair-wise pool comparison using the Agilent 2-color microarray platform. See Additional file 1: Figure S1 legend. (PDF 270 kb)
Additional file 5: Figure S5.
Bland-Altman plot of each pair-wise pool comparison using the LifeTech NGS platform. Symbols correspond to subpools A-E (see Fig. 1). Filled circles = A, open circles = B, open diamonds = C, open triangles = D, and open squares = E. (PDF 186 kb)
Additional file 6: Figure S6.
Bland-Altman plot of each pair-wise pool comparison using the Illumina NGS platform. See Additional file 5: Figure S5 legend. (PDF 172 kb)
Additional file 7:
Illumina bead ERCC controls data in comma-separated values (CSV) format. (CSV 15 kb)
Additional file 8:
NIAID microarray ERCC controls data in comma-separated values (CSV) format. (CSV 15 kb)
Additional file 9:
Agilent 1-color ERCC controls data in comma-separated values (CSV) format. (CSV 14 kb)
Additional file 10:
Agilent 2-color ERCC controls data in comma-separated values (CSV) format. (CSV 14 kb)
Additional file 11:
LifeTech NGS ERCC controls data in comma-separated values (CSV) format. (CSV 7 kb)
Additional file 12:
Illumina NGS ERCC controls data in comma-separated values (CSV) format. (CSV 9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pine, P.S., Munro, S.A., Parsons, J.R. et al. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design. BMC Biotechnol 16, 54 (2016). https://doi.org/10.1186/s12896-016-0281-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12896-016-0281-x